MIMOSA2
What Does MIMOSA2 Do? Description and Terminology
Input Data
Choosing Settings and Running the Analysis
Results
R Package Tutorial
Reference Data
FAQs
Citation and Contact
Using the mimosa2 R package
The easiest way to run a MIMOSA2 analysis is via the web application. However, you can also install the mimosa2 R package to run your own custom analyses and integrate MIMOSA2 into your analysis pipelines.
Installation
mimosa2 can be easily installed from GitHub using the devtools package:
devtools::install_github("borenstein-lab/mimosa2", dependencies = T)
library(mimosa)
If you see some warnings when loading the package, this is normal.
If you want to analyze ASV data, you will also need to have the program vsearch installed. Visit the vsearch website to download and install.
If you want to test your installation (including optionally access to vsearch), you can run:
test_m2_analysis(test_vsearch = T)
This will only test the package installation, not the setup of your reference data (see below).
Generating or downloading preprocessed reference data
Before running a MIMOSA2 analysis, the reference data you would like to use for the analysis needs to be downloaded and/or generated. MIMOSA2 relies on two separate types of reference data:
1) Reference data to link ASVs to reference taxa (not necessary if you have metagenomic KO annotation data)
2) Sets of genes and reactions linked to reference taxa
The figure below illustrates all the possible combinations of input and reference data formats. You can set up the reference databases to run all of them or just a subset for a specific analysis.
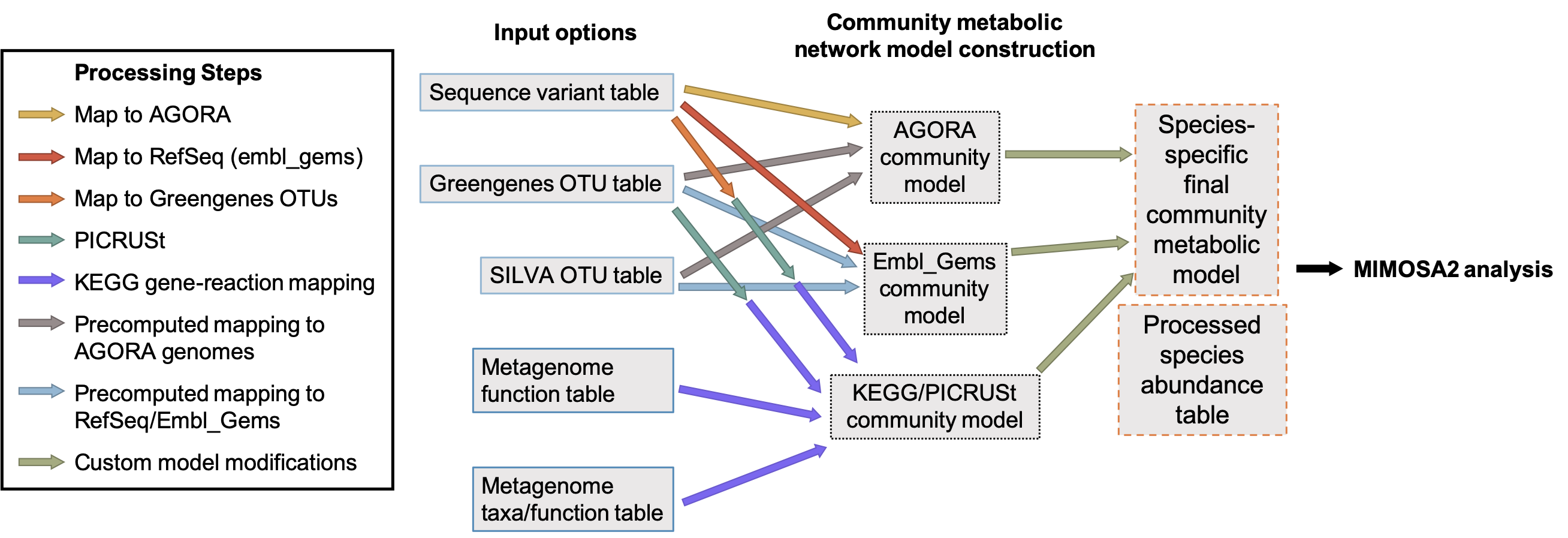
Several of these are available from the Downloads page. These can also be regenerated using scripts provided in the MIMOSA2 GitHub repository.
If you would like to run a workflow that uses freely available data, the download_reference_data function will obtain the necessary data and format it as expected by the main MIMOSA2 analysis. This function takes two arguments, which correspond to the file1_type and ref_choices options in the configuration table for a MIMOSA2 analysis (below).
download_reference_data(seq_db = "Sequence variants (ASVs)", target_db = "AGORA genomes and models")
download_reference_data(seq_db = "Greengenes 13_5 or 13_8 OTUs", target_db = "RefSeq/EMBL_GEMs genomes and models")
You can use the save_to argument to customize where these files are saved, but if you change this you will need to modify the data_prefix argument when running your MIMOSA2 analysis (see below). The result of this function should be to produce a directory called “data” containing a sub-directory called either “AGORA” or “embl_gems”, which contains mapping data as well as a further subdirectory called “RxnNetworks” containing metabolic reference data for each taxon.
KEGG setup
If you would like to run an analysis using KEGG, you need to have a KEGG license and to download 3 files from the KEGG FTP server: annotated pathway reactions (filename reaction_mapformula.lst), reaction annotations (filename reaction), and reaction-KO links (filename ko_reaction.list). Then you can provide those files as input to the generate_preprocessed_networks function to set up the reference database for MIMOSA2. For example:
generate_preprocessed_networks(database = "KEGG", kegg_paths = c("~/Downloads/reaction_mapformula.lst", "~/Downloads/reaction_ko.list", "~/Downloads/reaction"), out_path = "MIMOSA2_analysis/data/KEGG/")
It is not a problem if this command generates a few warnings.
Run a full MIMOSA2 analysis
Once you have downloaded and set up the relevant reference databases, you can run a full MIMOSA2 analysis simply by providing a “configuration table” containing all of the relevant settings for the analysis to the run_mimosa2 function.
The table below lists the various fields that you can provide in your configuration table. Required fields are in bold. The fields or rows of the table can be specified in any order.
| Field | Description | Possible values |
|---|---|---|
| file1 | Microbiome file path | Valid file path |
| file2 | Metabolomics file path | Valid file path |
| file1_type | Taxonomic abundance file type | One of: “Sequence variants (ASVs)”, “Greengenes 13_5 or 13_8 OTUs”, “SILVA 132 OTUs”, “Metagenome: Total KO abundances”, “Metagenome: Taxon-stratified KO abundances (HUMAnN2 or PICRUSt/PICRUSt2)” |
| ref_choices | Ref model option | One of: “PICRUSt KO genomes and KEGG metabolic model”, “AGORA genomes and models”, “RefSeq/EMBL_GEMs genomes and models” |
| data_prefix | File path to reference databases (see below for required files) | Valid file path |
| simThreshold | If 16S rRNA ASVs are provided, threshold for mapping them to a reference database | Value from 0 to 1 (default 0.99) |
| netAdd | File path to network modifications file | Valid file path |
| metType | Whether metabolite data is provided as KEGG compound IDs or metabolite names (assumes KEGG if not provided) | One of: “KEGG Compound IDs”, “Metabolite names (search for matching ID)” (default “KEGG Compound IDs”) |
| signifThreshold | Taxonomic contributors to metabolites will only be evaluated for metabolites with a model fit p-value below this threshold | Value from 0 to 1 (default 0.2) |
| compare_only | Skip the taxonomic contribution analysis, only build the model and compare CMP scores and metabolites | T or F (default F) |
| logTransform | Whether a log transform should be applied to metabolite data | T or F (default F) |
| rankBased | Whether to use rank-based regression for comparing CMP scores and metabolites | T or F (default T) |
| vsearch_path | File path to vsearch executable | Valid path (when not provided, MIMOSA2 assumes vsearch is in the executable path) |
Some example configuration tables are linked below:
- An ASV-based analysis using EMBL_GEMS models and rank-based regression
- A stratified metagenome-based analysis using KEGG and OLS regression, with a custom network modification
- A GreenGenes OTU table mapped to AGORA models
You can also download the configuration table used to run any analysis on the MIMOSA2 web server, which allows anyone to later reproduce the same analysis in an R session.
Once you have downloaded the necessary reference data, installed MIMOSA2 and vsearch, and created a configuration table, it is easy to run a full MIMOSA2 analysis. Save it as a text document, for example “configuration_table1.txt”, and run the following in an R session or script:
mimosa_results = run_mimosa2("configuration_table1.txt")
The run_mimosa2 function returns a list of data tables that is identical to the set of results provided by the web application. More details about the results are provided on the Results page.
If you want to generate plots of metabolic potential and taxonomic contributors for each metabolite, similar to the web app, use the make_plots and save_plots arguments for run_mimosa2.
mimosa_results_make_plots = run_mimosa2("configuration_table1.txt", make_plots = T, save_plots = T)
In this case lists of plots will also be returned. If save_plots is true, the function will save all plots in a folder in the current working directory.
If you want to run MIMOSA2 using microbiome and metabolite data frames in an existing R session, you can use the species and mets arguments (below). Note that this means that the analysis will skip several initial data filters and quality checks that would otherwise be performed when importing the data.
mimosa_results = run_mimosa2("configuration_table1.txt", species = my_otu_table, mets = my_met_table)